2 Parsimony analysis
The phylogenetic dataset contains a high proportion of inapplicable codings (404/ 3325 = 12% of tokens), which are known to introduce error and bias to phylogenetic reconstruction ((Maddison 1993; Brazeau et al. 2017a)). As such, phylogenetic search employed a new algorithm that correctly handles inapplicable data (Brazeau et al. 2017a). This algorithm is implemented in the MorphyLib C library (Brazeau et al. 2017b), and phylogenetic search was conducted using the R package TreeSearch v0.0.8 (Smith 2018).
Namacalathus is included in the matrix but has been excluded from analysis due to its potentially long branch, which is likely to mislead analysis.
2.1 Search parameters
Heuristic searches were conducted using the parsimony ratchet (Nixon 1999) under equal and implied weights (Goloboff 1997) with a variety of concavity constants. The consensus tree presented in the main manuscript represents a strict consensus of all trees that are most parsimonious under one or more of the concavity constants (k) 2, 3, 4.5, 7, 10.5, 16 and 24, an approach that is known to produce higher accuracy than equal weights at any fixed level of precision (Smith 2017).
2.2 Analysis
2.2.1 Load data
2.2.2 Generate starting tree
Dailyatia has been selected as an outgroup as camenellans have been interpreted as the earliest diverging members of the Brachiozoa (Skovsted et al. 2015a; Zhao et al. 2017).
nj.tree <- NJTree(my_data)
rooted.tree <- EnforceOutgroup(nj.tree, 'Dailyatia')
start.tree <- TreeSearch(tree=rooted.tree, dataset=my_data, maxIter=3000,
EdgeSwapper=RootedNNISwap, verbosity=0)2.2.3 Implied weights analysis
for (k in kValues) {
iw.tree <- IWRatchet(start.tree, iw_data, concavity=k,
ratchHits = 60, searchHits=55,
swappers=list(RootedTBRSwap, RootedSPRSwap, RootedNNISwap),
verbosity=0)
score <- IWScore(iw.tree, iw_data, concavity=k)
# Write single best tree
write.nexus(iw.tree, file=paste0("TreeSearch/hy_iw_k", k, "_", signif(score, 5), ".nex", collapse=''))
suboptFraction = 0.02
iw.consensus <- IWRatchetConsensus(iw.tree, iw_data, concavity=k,
swappers=list(RootedTBRSwap, RootedNNISwap),
searchHits=4,
suboptimal=score * suboptFraction,
nSearch=150, verbosity=0L)
write.nexus(iw.consensus, file=paste0("TreeSearch/hy_iw_k", k, "_", signif(IWScore(iw.tree, iw_data, concavity=k), 5), ".all.nex", collapse=''))
}2.2.4 Equal weights analysis
ew.tree <- Ratchet(start.tree, my_data, verbosity=0L,
ratchHits = 10, searchHits=55,
swappers=list(RootedTBRSwap, RootedSPRSwap, RootedNNISwap))
write.nexus(best.tree, file=paste0("TreeSearch/hy_ew_", Fitch(ew.tree, my_data), ".nex", collapse=''))
ew.consensus <- RatchetConsensus(ew.tree, my_data, nSearch=150,
swappers=list(RootedTBRSwap, RootedNNISwap),
verbosity=0L)
write.nexus(ew.consensus, file=paste0("TreeSearch/hy_ew_", Fitch(ew.tree, my_data), ".nex", collapse=''))2.3 Results
2.3.1 Implied weights results
# Read results from files
iw.trees <- lapply(kValues, function (k) {
iw.best <- list.files('TreeSearch',
pattern=paste0('hy_iw_k',
gsub('\\.', '\\\\.', k),
'_\\d+\\.?\\d*\\.all\\.nex'),
full.names=TRUE)
# Return:
if (length(iw.best) == 0) {
list()
} else {
read.nexus(iw.best[which.max(file.mtime(iw.best))])
}
})omit=c(lon, 'Clupeafumosus_socialis',
'Heliomedusa_orienta', 'Haplophrentis_carinatus')
ColPlot(ConsensusWithout(lapply(iw.trees, consensus), omit))
ColMissing(omit)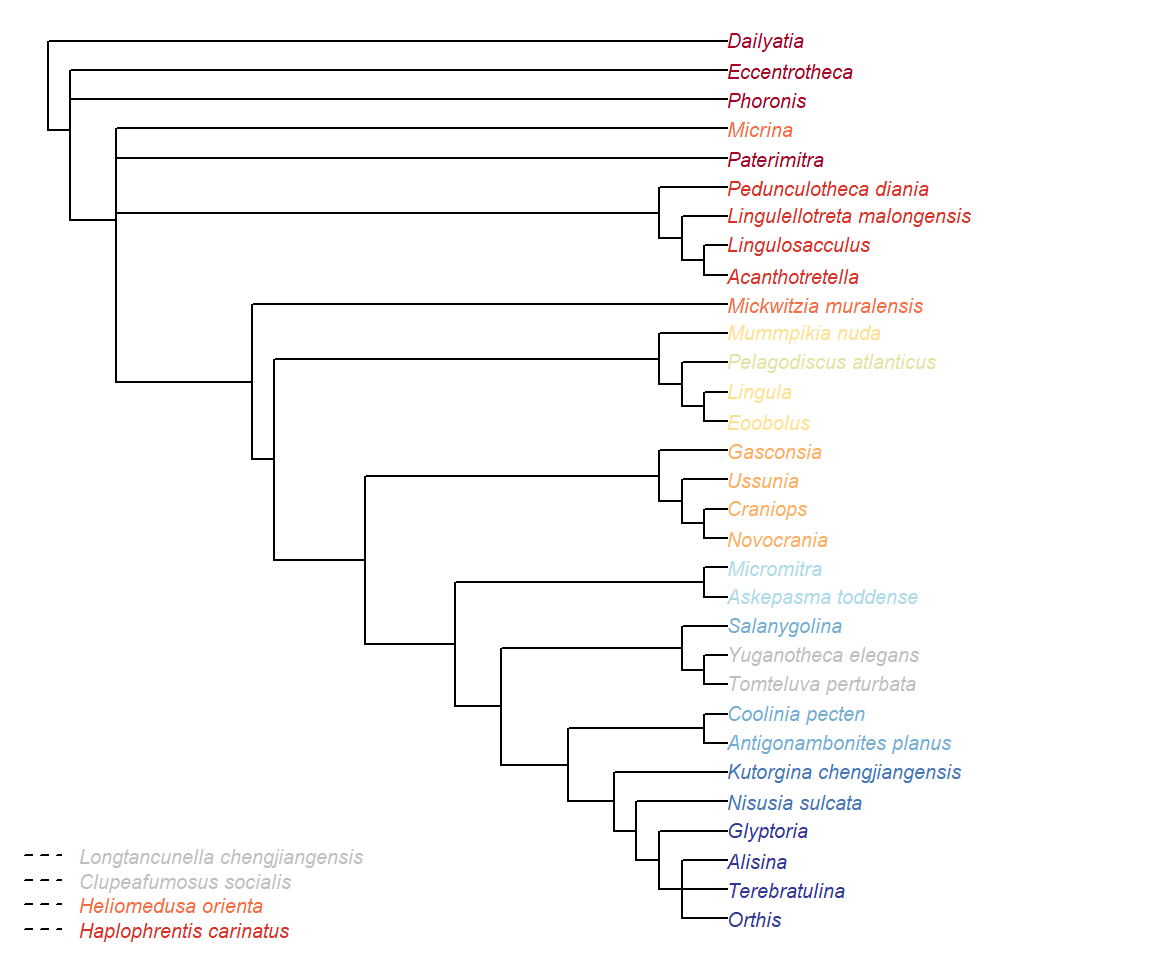
Figure 2.1: Consensus of implied weights analyses at all values of k
# Plot consensus results
par(mfrow=c(4, 2), mar=rep(0.2, 4))
ColPlot(consensus(lapply(iw.trees, consensus)))
text(-0.5, 1, pos=4, "Consensus of all k values", cex=0.8)
# Plot results for each value of k
for (i in seq_along(iw.trees)) {
ColPlot(consensus(iw.trees[[i]]))
text(1, 1, paste0('k = ', kValues[i]), pos=4)
}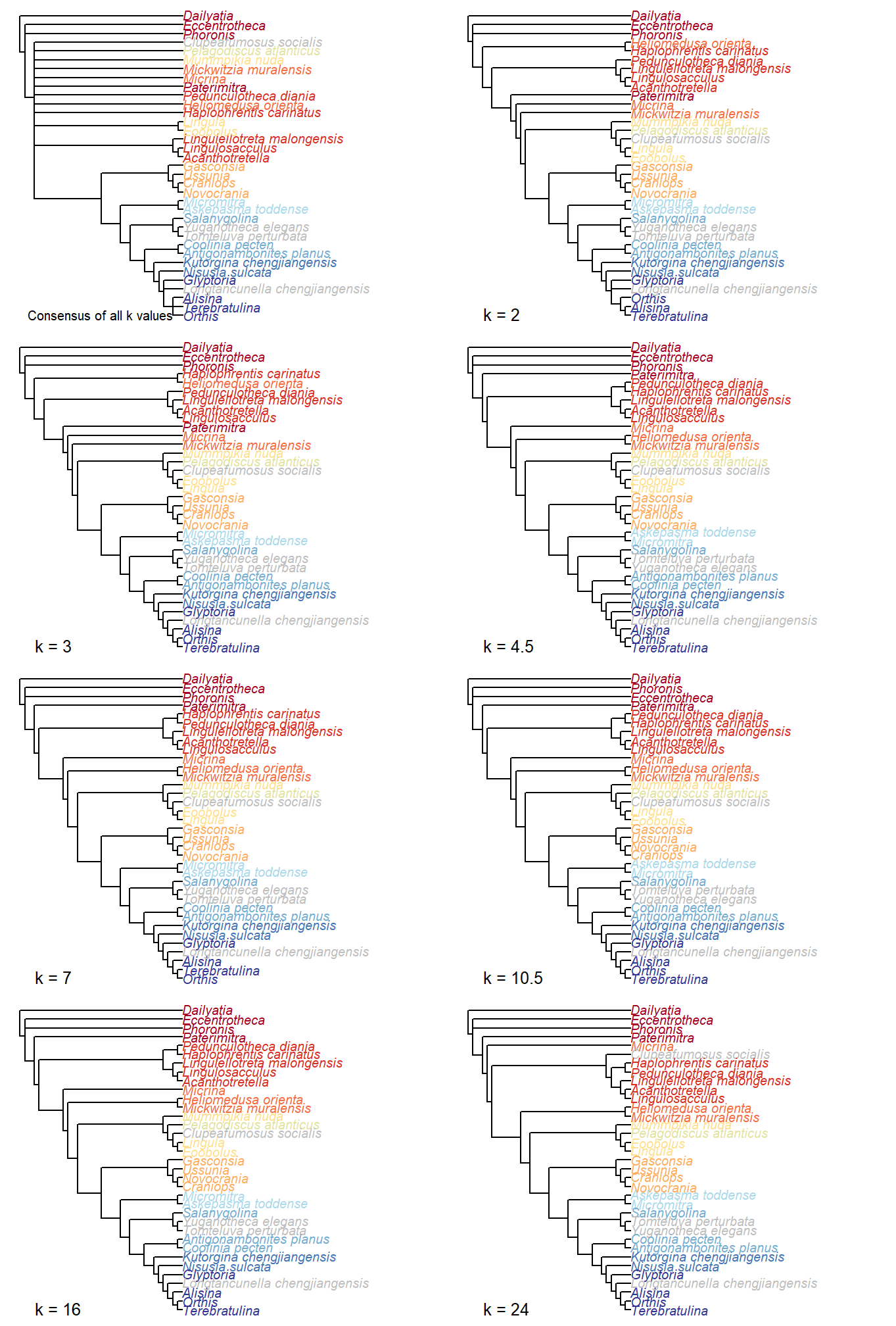
Figure 2.2: Implied weights results
2.3.2 Equal weights results
ew.best <- list.files('TreeSearch', pattern='hy_ew_\\d*\\.nex', full.names=TRUE)
ew.tree <- read.nexus(file=ew.best[which.max(file.mtime(ew.best))])
ColPlot(consensus(ew.tree))
(#fig:equal weights results in TreeSearch)Strict consensus of equal weights results
omit <- c(lon)
ColPlot(ConsensusWithout(ew.tree, omit))
ColMissing(omit)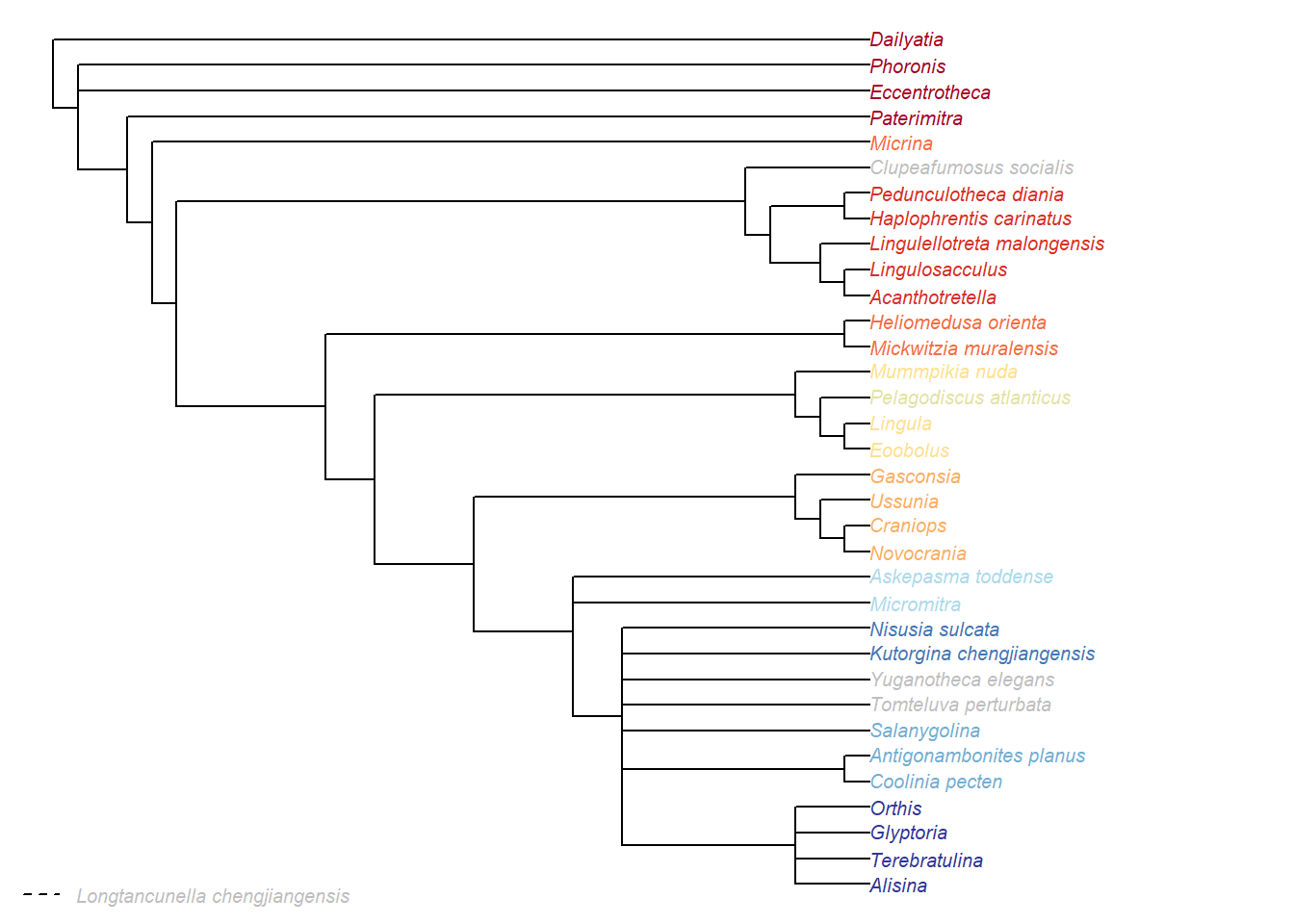
Figure 2.3: Strict consensus of equal weights results, taxa excluded
References
Maddison, W. P. 1993: Missing data versus missing characters in phylogenetic analysis. Systematic Biology 42:576–581.
Brazeau, M. D., T. Guillerme, and M. R. Smith. 2017a: Morphological phylogenetic analysis with inapplicable data. BioR\(\chi\)iv.
Brazeau, M. D., M. R. Smith, and T. Guillerme. 2017b: MorphyLib: a library for phylogenetic analysis of categorical trait data with inapplicability..
Smith, M. 2018: TreeSearch: phylogenetic tree search using custom optimality criteria..
Nixon, K. C. 1999: The Parsimony Ratchet, a new method for rapid parsimony analysis. Cladistics 15:407–414.
Goloboff, P. A. 1997: Self-weighted optimization: tree searches and character state reconstructions under implied transformation costs. Cladistics 13:225–245.
Smith, M. R. 2017: Quantifying and visualising divergence between pairs of phylogenetic trees: implications for phylogenetic reconstruction. BioR\(\chi\)iv.
Skovsted, C. B., M. J. Betts, T. P. Topper, and G. A. Brock. 2015a: The early Cambrian tommotiid genus Dailyatia from South Australia. Memoirs of the Association of Australasian Palaeontologists 48:1–117.
Zhao, F.-C., M. R. Smith, Z.-J. Yin, H. Zeng, G.-X. Li, and M.-Y. Zhu. 2017: Orthrozanclus elongata n. sp. and the significance of sclerite-covered taxa for early trochozoan evolution. Scientific Reports 7:16232.