Linear regression of life expectancy on year
Overview
We recently learned how to write our own R functions (Part 1, Part 2, Part 3).
Now we use that knowledge to write another useful function, within the context of the Gapminder data:
- Input: a data.frame that contains (at least) a life expectancy variable
lifeExpand a variable for yearyear - Output: a vector of estimated intercept and slope, from a linear regression of
lifeExponyear
The ultimate goal is to apply this function to the Gapminder data for a specific country. We will eventually scale up to all countries using external machinery, e.g., the dplyr::group_by() + dplyr::do().
Load the Gapminder data
As usual, load the Gapminder excerpt. Load ggplot2 because we’ll make some plots and dplyr too.
library(gapminder)
library(ggplot2)
suppressPackageStartupMessages(library(dplyr))Get data to practice with
I extract the data for one country in order to develop some working code interactively.
j_country <- "France" # pick, but do not hard wire, an example
(j_dat <- gapminder %>%
filter(country == j_country))
## country continent year lifeExp pop gdpPercap
## 1 France Europe 1952 67.410 42459667 7029.809
## 2 France Europe 1957 68.930 44310863 8662.835
## 3 France Europe 1962 70.510 47124000 10560.486
## 4 France Europe 1967 71.550 49569000 12999.918
## 5 France Europe 1972 72.380 51732000 16107.192
## 6 France Europe 1977 73.830 53165019 18292.635
## 7 France Europe 1982 74.890 54433565 20293.897
## 8 France Europe 1987 76.340 55630100 22066.442
## 9 France Europe 1992 77.460 57374179 24703.796
## 10 France Europe 1997 78.640 58623428 25889.785
## 11 France Europe 2002 79.590 59925035 28926.032
## 12 France Europe 2007 80.657 61083916 30470.017Always always always plot the data. Yes, even now.
p <- ggplot(j_dat, aes(x = year, y = lifeExp))
p + geom_point() + geom_smooth(method = "lm", se = FALSE)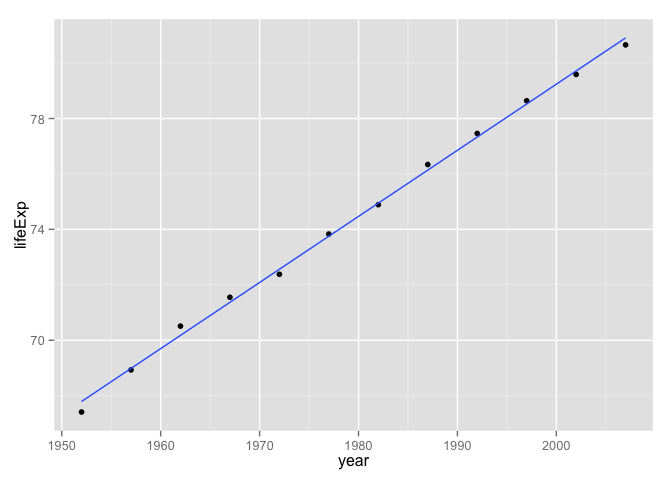
Get some code that works
Fit the regression
j_fit <- lm(lifeExp ~ year, j_dat)
coef(j_fit)
## (Intercept) year
## -397.7646019 0.2385014Whoa, check out that crazy intercept! Apparently the life expectancy in France around year 0 A.D. was minus 400 years! Never forget to sanity check a model. In this case, a reparametrization is in order. I think it makes more sense for the intercept to correspond to life expectancy in 1952, the earliest date in our dataset. Estimate the intercept eye-ball-o-metrically from the plot and confirm that we’ve got something sane and interpretable now.
j_fit <- lm(lifeExp ~ I(year - 1952), j_dat)
coef(j_fit)
## (Intercept) I(year - 1952)
## 67.7901282 0.2385014Turn working code into a function
Create the basic definition of a function and drop your working code inside. Add arguments and edit the inner code to match. Apply it to the practice data. Do you get the same result as before?
le_lin_fit <- function(dat, offset = 1952) {
the_fit <- lm(lifeExp ~ I(year - offset), dat)
coef(the_fit)
}
le_lin_fit(j_dat)
## (Intercept) I(year - offset)
## 67.7901282 0.2385014I had to decide how to handle the offset. Given that I will scale this up to many countries, which, in theory, might have data for different dates, I chose to set a default of 1952. Strategies that compute the offset from data, either the main Gapminder dataset or the excerpt passed to this function, are also reasonable to consider.
I loathe the names on this return value. This is not my first rodeo and I know that, downstream, these will contaminate variable names and factor levels and show up in public places like plots and tables. Fix names early!
le_lin_fit <- function(dat, offset = 1952) {
the_fit <- lm(lifeExp ~ I(year - offset), dat)
setNames(coef(the_fit), c("intercept", "slope"))
}
le_lin_fit(j_dat)
## intercept slope
## 67.7901282 0.2385014Much better!
Test on other data and in a clean workspace
It’s always good to rotate through examples during development. The most common error this will help you catch is when you accidentally hard-wire your example into your function. If you’re paying attention to your informal tests, you will find it creepy that your function returns exactly the same results regardless which input data you give it. This actually happened to me while I was writing this document, I kid you not! I had left j_fit inside the call to coef(), instead of switching it to the_fit. How did I catch that error? I saw the fitted line below, which clearly did not have an intercept in the late 60s and a positive slope, as my first example did. Figures are a mighty weapon in the fight against nonsense. I don’t trust analyses that have few/no figures.
j_country <- "Zimbabwe"
(j_dat <- gapminder %>% filter(country == j_country))
## country continent year lifeExp pop gdpPercap
## 1 Zimbabwe Africa 1952 48.451 3080907 406.8841
## 2 Zimbabwe Africa 1957 50.469 3646340 518.7643
## 3 Zimbabwe Africa 1962 52.358 4277736 527.2722
## 4 Zimbabwe Africa 1967 53.995 4995432 569.7951
## 5 Zimbabwe Africa 1972 55.635 5861135 799.3622
## 6 Zimbabwe Africa 1977 57.674 6642107 685.5877
## 7 Zimbabwe Africa 1982 60.363 7636524 788.8550
## 8 Zimbabwe Africa 1987 62.351 9216418 706.1573
## 9 Zimbabwe Africa 1992 60.377 10704340 693.4208
## 10 Zimbabwe Africa 1997 46.809 11404948 792.4500
## 11 Zimbabwe Africa 2002 39.989 11926563 672.0386
## 12 Zimbabwe Africa 2007 43.487 12311143 469.7093
p <- ggplot(j_dat, aes(x = year, y = lifeExp))
p + geom_point() + geom_smooth(method = "lm", se = FALSE)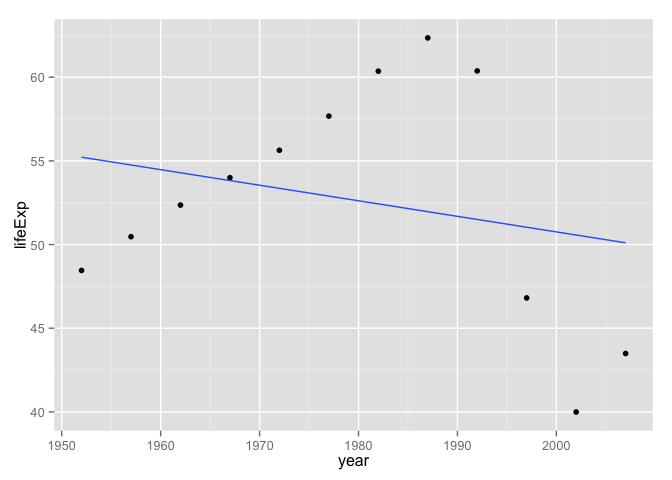
le_lin_fit(j_dat)
## intercept slope
## 55.22124359 -0.09302098The linear fit is comically bad, but yes I believe the visual line and the regression results match up.
It’s also a good idea to clean out the workspace, rerun the minimum amount of code, and retest your function. This will help you catch another common mistake: accidentally relying on objects that were lying around in the workspace during development but that are not actually defined in your function nor passed as formal arguments.
rm(list = ls())
le_lin_fit <- function(dat, offset = 1952) {
the_fit <- lm(lifeExp ~ I(year - offset), dat)
setNames(coef(the_fit), c("intercept", "slope"))
}
le_lin_fit(gapminder %>% filter(country == "Zimbabwe"))
## intercept slope
## 55.22124359 -0.09302098Are we there yet?
Yes.
Given how I plan to use this function, I don’t feel the need to put it under formal unit tests or put in argument validity checks. Let’s move on to the exciting part, which is scaling this up to all countries.
