Made for bioinformatics and life sciences, MOLGENIS compute is a flexible shell script framework to generate big data workflows that can run parallel on clusters and grids. Molgenis Compute Users can:
- Design a workflow.csv with each step as a shell script protocol;
- Generate and run jobs by iterating over parameters.csv and execute on a compute backend;
- (Optional) use standardized file and tool management for portable workflows.
2. Get Started
The latest distribution of Molgenis Compute is available at the Molgenis Compute start page. Unzip the archive and the command-line version is ready to use.
unzip molgenis-compute-core-0.0.1-SNAPSHOT-distribution.zip cd molgenis-compute-core-0.0.1-SNAPSHOT
3. Create workflow
Now u can create your first workflow by executing the following command
sh molgenis_compute.sh --create myfirst_workflow
Afterwards go to the created workflow directory.
cd myfirst_workflow
You see the typical Molgenis Compute workflow structure
/protocols #folder with bash script 'protocols' /protocols/step1.sh #example of a protocol shell script /protocols/step2.sh #example of a protocol shell script workflow.csv #file listing steps and parameter flow workflow.defaults.csv #default parameters for workflow.csv (optional) parameters.csv #parameters you want to run analysis on header.ftl #user extra script header (optinal) footer.ftl #user extra script footer (optinal)
A similar structure should be created for every workflow. Additionally, a workflow can be read from the github repository, if the github root is specified.
In the easiest scenario, the
workflow.csv file has the following structure:
step,protocol,dependencies step1,protocols/step1.sh, step2,protocols/step2.sh,step1
This means that the workflow consists of two steps step1 and step2, where step2 depends on step1.
step1 has its analysis protocol in the file protocols/step1.sh and step2 in the file protocols/step2.sh respectively.
The created workflow in our example workflow.csv is a bit more complex.
Let’s first look at the parameters.csv file, which contains some workflow parameters. In this example, one parameter
input has two values hello and bye.
input hello bye
These parameters can be used in protocols. In the following protocol example
#string input
#output out
# Let's do something with string 'input'
echo ${input}_hasBeenInStep1
out=${input}_hasBeenInStep1
input will be substituted with values hello or bye. In the header of protocols, we specify inputs with flags #string for variables with a single value and #list for variables with multiple values. The outputs are specified with the flag #output
In our example protocol step1.sh , we would like to call input as in
step1.ftl
#string in
#output out
# Let's do something with string 'in'
echo ${in}_hasBeenInStep1
out=${in}_hasBeenInStep1
In this case, we need to map these names in our example workflow.csv file
step,protocol,parameterMapping step1,protocols/step1.sh,in=input step2,protocols/step2.sh,wf=workflowName;date=creationDate;strings=step1.out
in=input
This does the trick. In the same way, we can map outputs of one step to the inputs of the next steps. In our example, strings in the step2, which has protocol
step2.ftl
#string wf
#string date
#list strings
echo "Workflow name: ${wf}"
echo "Created: ${date}"
echo "Result of step1.sh:"
for s in "${strings[@]}"
do
echo ${s}
done
echo "(FOR TESTING PURPOSES: your runid is ${runid})"
are mapped using
strings=step1.out
Here, prefix step1. says that out is coming from step1.
The example protocols has the following listings:
In our example variables date and wf are defined in
an additional parameters file workflow.defaults.csv.
workflowName,creationDate myFirstWorkflow,today
In this way, the parameters can be divided in several groups and re-used in different workflows. If users do not like to map parameters, they should use the same names in protocols and parameters files. This makes parameters a kind of global.
4. Generate workflow
To generate actual workflow jobs, run the next command-line
sh molgenis_compute.sh --generate --parameters myfirst_workflow/parameters.csv --workflow myfirst_workflow/workflow.csv --defaults myfirst_workflow/workflow.defaults.csv
or with a short command-line version
sh molgenis_compute.sh -g -p myfirst_workflow/parameters.csv -w myfirst_workflow/workflow.csv -defaults myfirst_workflow/workflow.defaults.csv
The directory rundir is created.
ls rundir/
It contains a number of files
doc step1_0.sh step1_1.sh step2_0.sh submit.sh user.env
.sh are actual scripts generated from the specified workflow. step1 has two scripts and step2 has only one, because it treats
outputs from scripts of the step1 as a list, which is specified in step2.sh by
#list strings
user.env contains all actual parameters mappings. In this example:
# ## User parameters # creationDate[0]="today" creationDate[1]="today" input[0]="hello" input[1]="bye" workflowName[0]="myFirstWorkflow" workflowName[1]="myFirstWorkflow"
Parameters, which are known before hand can be connected to the environment file or weaved directly in the protocols (if weave flag is set in command-line options). In our example, two shell scripts are generated for the step1. The weaved version of generated files are shown below.
step1_0.sh
#string in #output out # Let's do something with string 'in' echo "hello_hasBeenInStep1" out=hello_hasBeenInStep1
and
step1_1.sh
#string in #output out # Let's do something with string 'in' echo "bye_hasBeenInStep1" out=bye_hasBeenInStep1
The output values of the first steps are not known beforehand, so, string cannot be weaved and will stay in the generated for the step2 script as it was. However, the wf and date values are weaved.
step2_0.sh
#string wf
#string date
#list strings
echo "Workflow name: myFirstWorkflow"
echo "Created: today"
echo "Result of step1.sh:"
for s in "${strings[@]}"
do
echo ${s}
done
If values can be known, the script will have the following content
step2_0.sh with all known values
#string wf
#string date
#list strings
echo "Workflow name: myFirstWorkflow"
echo "Created: today"
echo "Result of step1.sh:"
for s in "hello" "bye"
do
echo ${s}
done
If weaved flag is not set, step1_0.sh file, for example looks as follows:
# Connect parameters to environment
input="bye"
#string input
# Let's do something with string 'in'
echo "${input}_hasBeenInStep1"
out=${input}_hasBeenInStep1
In this way, users can choose how generated files look like. In the current implementation, values are first taken from parameter files. If they are not present, then compute looks, if these values can be known at run-time, by analysing all previous steps of the protocol, where values are unknown. If values cannot be known at run-time, compute will give a generation error.
5. Execute workflow
The workflow can be executed with the command
sh molgenis_compute.sh --run ls rundir/
Now, rundir contains more files
doc step1_0.sh.finished step1_1.sh.finished step2_0.sh.finished molgenis.bookkeeping.log step1_0.sh.started step1_1.sh.started step2_0.sh.started step1_0.env step1_1.env step2_0.env submit.sh step1_0.sh step1_1.sh step2_0.sh user.env
.started and .finished files are created, when certain jobs are started and finished respectively.
In our example, strings variable from step2 requires run-time values produced in step1. These values are taken from
step1_X.env files. For example:
step1_0.env
step1__has__out[0]=hello_hasBeenInStep1
In the workflow file, it is specified with a simple .
strings=step1.out
and substituted with has in generated script files.
6. Command-line options
Molgenis Compute has the following command-line options:
### MOLGENIS COMPUTE ###
Version: development
usage: sh molgenis-compute.sh -p parameters.csv
-b,--backend <arg> Backend for which you generate.
Default: localhost
-bp,--backendpassword <arg> Supply user pass to login to execution
backend. Default is not saved.
-bu,--backenduser <arg> Supply user name to login to execution
backend. Default is your own user
name.
-clear,--clear Clear properties file
-create <arg> Creates empty workflow. Default name:
myworkflow
-d,--database <arg> Host, location of database. Default:
none
-dbe,--database-end End the database
-dbs,--database-start Starts the database
-defaults,--defaults <arg> Path to your workflow-defaults file.
Default: defaults.csv
-footer <arg> Adds a custom footer. Default:
footer.ftl
-g,--generate Generate jobs
-h,--help Shows this help.
-header <arg> Adds a custom header. Default:
header.ftl
-l,--list List jobs, generated, queued, running,
completed, failed
-mpass,--molgenispassword <arg> Supply user pass to login to molgenis.
Default is not saved.
-mu,--molgenisuser <arg> Supply user name to login to molgenis
database. Default is your own user
name.
-o,--overwrite <arg> Parameters and values, which will
overwritten in the parameters file.
Parameters should be placed into
brackets and listed using equality
sign, e.g. "mem=6GB;queue=long"
-p,--parameters <parameters.csv> Path to parameter.csv file(s).
Default: parameters.csv
-path,--path Path to directory this generates to.
Default: <current dir>.
-port,--port <arg> Port used to connect to databasae.
Default: 8080
-r,--run Run jobs from current directory on
current backend. When using --database
this will return a 'id' for --pilot.
-rundir <arg> Directory where jobs are stored
-runid,--runid <arg> Id of the task set which you generate.
Default: null
-submit <arg> Set a custom submit.sh template.
Default: submit.sh.ftl
-w,--workflow <workflow.csv> Path to your workflow file. Default:
workflow.csv
-weave,--weave Weave parameters to the actual script
-web,--web <arg> Location of the workflow in the public
github repository. The other
parameters should be specified
relatively to specified github root.
7. Reserved words
Molgenis Compute has a list of reserved words, which cannot be used in compute to name parameters, workflow steps, etc. These words are listed below:
port interval workflow path defaults parameters rundir runid backend database walltime nodes ppn queue mem _NA password molgenisuser backenduser header footer submit autoid
The reserved words are used in the compute.properties file. This file is created to save the latest compute configuration and discuss further.
8. Advanced parameter formats
More parameters can be specified using the next format
parameter1, parameter2 value11, value21 value12, value22 value13, value23
Alternatively, parameters can be specified in the .properties style. The parameters file also should have
the .properties extension.
parameter1 = value11, value12, value13 parameter2 = value21, value22, value23
Values for the workflow to iterate over can be passed as CSV file with parameter names in the header (a-zA-Z0-9 and underscore, starting with a-zA-Z) and parameter values in each row. Use quotes to escape commas, e.g. "a,b".
Each value is one of the following:
- a string
- a Freemarker template Freemarker
- a series i..j (where i and j are two integers), meaning from i to j including i and j
- a list of ; separated values (may contain templates)
You can combine multiple parameter files: the values will be natural joined based on overlapping columns.
Example with two or more parameter files:
molgenis --path path/to/workflow -p f1.csv -p f2.csv -w workflow.csv
f1.csv (white space will be trimmed):
p0, p2 x, 1 y, 2
f2.csv (white space will be trimmed):
p1, p2, p3, p4
v1, 1..2, a;b, file${p2}
Merged and expanded result for f1.csv + f2.csv:
p0, p1, p2, p3, p4 x, v1, 1, a, file1 x, v1, 1, b, file1 y, v1, 2, a, file2 y, v1, 2, b, file2
More complex parameter examples can combine values with template, as following:
foo= item1 , item2
bar= ${foo}, item3
number= 123
Here, variable bar has two values of variable foo.
Specifying workflow in parameters file
Alternatively to specifying workflow in the command-line using -w or --workflow, workflow can be present as a parameter in
parameters.csv file:
workflow, parameter1, parameter2 workflow.csv, value1, value2
Lists of parameters
Parameters can be specified in several parameter files. To understand how list parameter specification works, let’s consider the case with 2 parameter files and 1 protocol.
parameters1.csv
project , sample project1, sample1 project1, sample2 project1, sample3
parameters2.csv
chr chr1 chr2 chr3
The example protocol looks like
protocol1.sh
#!/bin/sh
#string project
#list sample
#list chr
for s in "${sample[@]}"
do
echo $s
for c in "${chr[@]}"
do
echo $c
done
done
Here, sample and chr parameters are coming from 2 different parameter files. Both parameters are specified as list in the protocol. These lists of parameters will not be combined, since they are coming from different parameters files. The generated parameters lists will have the next look:
#!/bin/sh
#string project
#list sample
#list chr
for s in "sample1" "sample2" "sample3"
do
echo $s
for c in "chr1" "chr2" "chr3"
do
echo $c
done
done
If users want to combine lists that coming from separated files, lists should be declared on the same line, like
list sample, chr
It will produce one list with all possible combination of parameters:
sample1, chr1 sample1, chr2 sample1, chr3 sample2, chr1 sample2, chr2 sample2, chr3 sample3, chr1 sample3, chr2 sample3, chr3
It is not the desired behaviour in the considered protocol:
#!/bin/sh
#string project
#list sample, chr
for s in "sample1" "sample1" "sample1" "sample2" "sample2" "sample2" "sample3" "sample3" "sample3"
do
echo $s
for c in "chr1" "chr2" "chr3" "chr1" "chr2" "chr3" "chr1" "chr2" "chr3"
do
echo $c
done
done
9. Script generation for PBS cluster and other back-ends
To generate for pbs, the next options should be added to the command line
--backend pbs
When generating script for computational clusters or grid, some additional parameters, such as execution wall-time, memory requirement, etc. should be specified.
This can be done in the parameters file
workflowName,creationDate,queue,mem,walltime,nodes,ppn myFirstWorkflow,today,short_queue,4GB,05:59:00,1,1
queue - cluster/grid queue mem - memory required walltime - execution wall time nodes - number of nodes needed ppn - number of cores needed
Or also it can be specified in the molgenis header in protocols
step1.ftl with molgenis header
#MOLGENIS queue=short_queue mem=4gb walltime=05:59:00 nodes=1 ppn=1
#string in
#output out
# Let's do something with string 'in'
out=${in}_hasBeenInStep1
The specification in protocols has priority over specification in parameter files.
In the command-line distribution, users can add a new back-end by adding a new directory, that contains header/footer and submit templates for that backend.
10. Switching to a different workflow
It is very advisable to start working with a new workflow with running
sh molgenis_compute.sh --clear
This command clears the .compute.properties file, which contains previous generation and running options.
11. Commenting in workflow specification
User can want to run only one or several steps of the workflow, when the rest of workflow can be commented out using # sign. In this example step2 is commented out.
step,protocol,dependencies step1,protocols/step1.sh, #step2,protocols/step2.sh,step1
12. Database usage
The detailed user gid for molgenis-compute database version will be added soon. The application can be build as a maven project. During the first start of compute-db, the admin with password admin will be created. So, the users can login into the system for the first time.
To insert new compute runs into database, use the command-line like
sh molgenis_compute.sh \ --generate \ --workflow myfirst_workflow/workflow.csv \ --defaults myfirst_workflow/workflow.defaults.csv \ --parameters myfirst_workflow/parameters.csv \ --rundir run \ --runid demo1 \ --database <db-ip-address> \ -mu <db-username> -mpass <db-password> \ -b <back-end> \ -bu <back-end-username> -bp <back-end-password>
After this your compute run will appear at the Jobs Dashboard. It will look like in the image below: User can submit jobs entering the user name and password for computational backend.
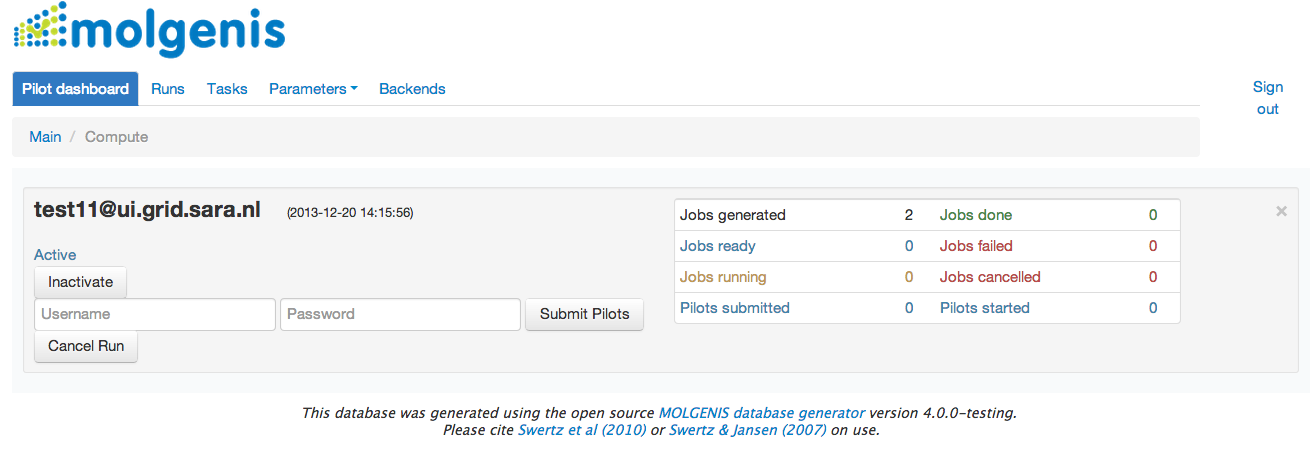
The jobs details can be expected in Tasks/History generated forms Jobs also can be submitted and monitored from the command-line using --run flag. However, this functionality is not fully tested yet and submission via web-user interface is more stable.
13. Available workflows
The available (NGS alignment, imputation) workflows are available in the workflow github repository.