Resampling
Resampling strategies are usually used to assess the performance of a learning algorithm: The entire data set is (repeatedly) split into training sets and test sets , . The learner is trained on each training set, predictions are made on the corresponding test set (sometimes on the training set as well) and the performance measure is calculated. Then the individual performance values are aggregated, most often by calculating the mean. There exist various different resampling strategies, for example cross-validation and bootstrap, to mention just two popular approaches.
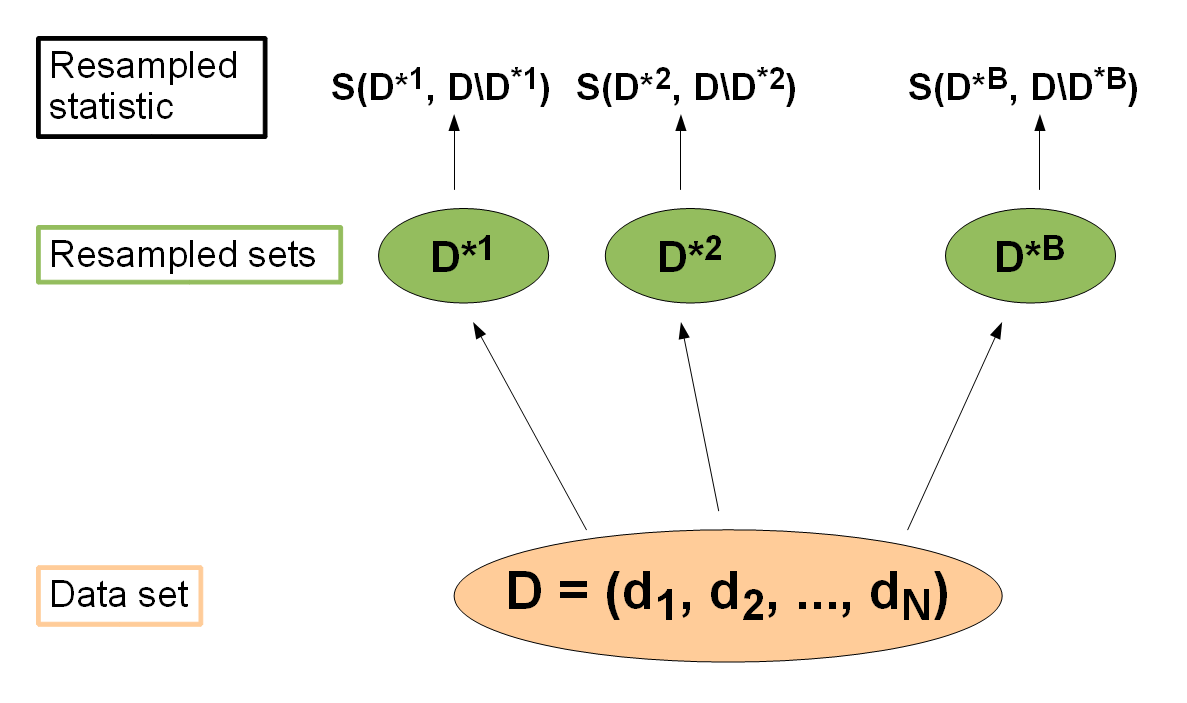
If you want to read up on further details, the paper Resampling Strategies for Model Assessment and Selection by Simon is probably not a bad choice. Bernd has also published a paper Resampling methods for meta-model validation with recommendations for evolutionary computation which contains detailed descriptions and lots of statistical background information on resampling methods.
Defining the resampling strategy
In mlr the resampling strategy can be defined via function makeResampleDesc. It requires a string that specifies the resampling method and, depending on the selected strategy, further information like the number of iterations. The supported resampling strategies are:
- Cross-validation (
"CV"), - Leave-one-out cross-validation (
"LOO"), - Repeated cross-validation (
"RepCV"), - Out-of-bag bootstrap and other variants like b632 (
"Bootstrap"), - Subsampling, also called Monte-Carlo cross-validation (
"Subsample"), - Holdout (training/test) (
"Holdout").
For example if you want to use 3-fold cross-validation type:
## 3-fold cross-validation
rdesc = makeResampleDesc("CV", iters = 3)
rdesc
#> Resample description: cross-validation with 3 iterations.
#> Predict: test
#> Stratification: FALSE
For holdout estimation use:
## Holdout estimation
rdesc = makeResampleDesc("Holdout")
rdesc
#> Resample description: holdout with 0.67 split rate.
#> Predict: test
#> Stratification: FALSE
In order to save you some typing mlr contains some pre-defined resample descriptions for very common strategies like holdout (hout) as well as cross-validation with different numbers of folds (e.g., cv5 or cv10).
hout
#> Resample description: holdout with 0.67 split rate.
#> Predict: test
#> Stratification: FALSE
cv3
#> Resample description: cross-validation with 3 iterations.
#> Predict: test
#> Stratification: FALSE
Performing the resampling
Function resample evaluates a Learner on a given machine learning Task using the selected resampling strategy.
As a first example, the performance of linear regression on the BostonHousing data set is calculated using 3-fold cross-validation.
Generally, for -fold cross-validation the data set is partitioned into subsets of (approximately) equal size. In the -th of the iterations, the -th subset is used for testing, while the union of the remaining parts forms the training set.
As usual, you can either pass a Learner object to resample or, as done here,
provide the class name "regr.lm" of the learner.
Since no performance measure is specified the default for regression learners
(mean squared error, mse) is calculated.
## Specify the resampling strategy (3-fold cross-validation)
rdesc = makeResampleDesc("CV", iters = 3)
## Calculate the performance
r = resample("regr.lm", bh.task, rdesc)
#> Resampling: cross-validation
#> Measures: mse
#> [Resample] iter 1: 19.8982806
#> [Resample] iter 2: 31.7803405
#> [Resample] iter 3: 20.9698523
#>
#> Aggregated Result: mse.test.mean=24.2161578
#>
r
#> Resample Result
#> Task: BostonHousing-example
#> Learner: regr.lm
#> Aggr perf: mse.test.mean=24.2161578
#> Runtime: 0.0628207
The result r is an object of class ResampleResult.
It contains performance results for the learner and some additional information
like the runtime, predicted values, and optionally the models fitted in single resampling
iterations.
## Peak into r
names(r)
#> [1] "learner.id" "task.id" "task.desc" "measures.train"
#> [5] "measures.test" "aggr" "pred" "models"
#> [9] "err.msgs" "err.dumps" "extract" "runtime"
r$aggr
#> mse.test.mean
#> 24.21616
r$measures.test
#> iter mse
#> 1 1 19.89828
#> 2 2 31.78034
#> 3 3 20.96985
r$measures.test gives the performance on each of the 3 test data sets.
r$aggr shows the aggregated performance value.
Its name "mse.test.mean" indicates the performance measure, mse,
and the method, test.mean, used to aggregate the 3 individual performances.
test.mean is the default aggregation scheme for most performance measures
and, as the name implies, takes the mean over the performances on the test data sets.
Resampling in mlr works the same way for all types of learning problems and learners. Below is a classification example where a classification tree (rpart) is evaluated on the Sonar data set by subsampling with 5 iterations.
In each subsampling iteration the data set is randomly partitioned into a training and a test set according to a given percentage, e.g., 2/3 training and 1/3 test set. If there is just one iteration, the strategy is commonly called holdout or test sample estimation.
You can calculate several measures at once by passing a list of Measures to resample. Below, the error rate (mmce), false positive and false negative rates (fpr, fnr), and the time it takes to train the learner (timetrain) are estimated by subsampling with 5 iterations.
## Subsampling with 5 iterations and default split ratio 2/3
rdesc = makeResampleDesc("Subsample", iters = 5)
## Subsampling with 5 iterations and 4/5 training data
rdesc = makeResampleDesc("Subsample", iters = 5, split = 4/5)
## Classification tree with information splitting criterion
lrn = makeLearner("classif.rpart", parms = list(split = "information"))
## Calculate the performance measures
r = resample(lrn, sonar.task, rdesc, measures = list(mmce, fpr, fnr, timetrain))
#> Resampling: subsampling
#> Measures: mmce fpr fnr timetrain
#> [Resample] iter 1: 0.2619048 0.2352941 0.2800000 0.0230000
#> [Resample] iter 2: 0.2857143 0.2857143 0.2857143 0.0190000
#> [Resample] iter 3: 0.2619048 0.2857143 0.2380952 0.0190000
#> [Resample] iter 4: 0.3333333 0.3500000 0.3181818 0.0200000
#> [Resample] iter 5: 0.3333333 0.1666667 0.4583333 0.0190000
#>
#> Aggregated Result: mmce.test.mean=0.2952381,fpr.test.mean=0.2646779,fnr.test.mean=0.3160649,timetrain.test.mean=0.0200000
#>
r
#> Resample Result
#> Task: Sonar-example
#> Learner: classif.rpart
#> Aggr perf: mmce.test.mean=0.2952381,fpr.test.mean=0.2646779,fnr.test.mean=0.3160649,timetrain.test.mean=0.0200000
#> Runtime: 0.178369
If you want to add further measures afterwards, use addRRMeasure.
## Add balanced error rate (ber) and time used to predict
addRRMeasure(r, list(ber, timepredict))
#> Resample Result
#> Task: Sonar-example
#> Learner: classif.rpart
#> Aggr perf: mmce.test.mean=0.2952381,fpr.test.mean=0.2646779,fnr.test.mean=0.3160649,timetrain.test.mean=0.0200000,ber.test.mean=0.2903714,timepredict.test.mean=0.0066000
#> Runtime: 0.178369
By default, resample prints progress messages and intermediate results. You can turn this off by setting
show.info = FALSE, as done in the code chunk below. (If you are interested in suppressing
these messages permanently have a look at the tutorial page about configuring mlr.)
In the above example, the Learner was explicitly constructed. For convenience
you can also specify the learner as a string and pass any learner parameters via the ... argument
of resample.
r = resample("classif.rpart", parms = list(split = "information"), sonar.task, rdesc,
measures = list(mmce, fpr, fnr, timetrain), show.info = FALSE)
r
#> Resample Result
#> Task: Sonar-example
#> Learner: classif.rpart
#> Aggr perf: mmce.test.mean=0.3047619,fpr.test.mean=0.2785319,fnr.test.mean=0.3093917,timetrain.test.mean=0.0202000
#> Runtime: 0.175945
Accessing resample results
Apart from the learner performance you can extract further information from the resample results, for example predicted values or the models fitted in individual resample iterations.
Predictions
Per default, the ResampleResult contains the predictions made during the resampling.
If you do not want to keep them, e.g., in order to conserve memory,
set keep.pred = FALSE when calling resample.
The predictions are stored in slot $pred of the resampling result, which can also be accessed
by function getRRPredictions.
r$pred
#> Resampled Prediction for:
#> Resample description: subsampling with 5 iterations and 0.80 split rate.
#> Predict: test
#> Stratification: FALSE
#> predict.type: response
#> threshold:
#> time (mean): 0.01
#> id truth response iter set
#> 1 36 R M 1 test
#> 2 132 M R 1 test
#> 3 145 M R 1 test
#> 4 161 M R 1 test
#> 5 108 M M 1 test
#> 6 178 M M 1 test
#> ... (#rows: 210, #cols: 5)
pred = getRRPredictions(r)
pred
#> Resampled Prediction for:
#> Resample description: subsampling with 5 iterations and 0.80 split rate.
#> Predict: test
#> Stratification: FALSE
#> predict.type: response
#> threshold:
#> time (mean): 0.01
#> id truth response iter set
#> 1 36 R M 1 test
#> 2 132 M R 1 test
#> 3 145 M R 1 test
#> 4 161 M R 1 test
#> 5 108 M M 1 test
#> 6 178 M M 1 test
#> ... (#rows: 210, #cols: 5)
pred is an object of class ResamplePrediction.
Just as a Prediction object (see the tutorial page on making predictions)
it has an element $data which is a data.frame that contains the
predictions and in the case of a supervised learning problem the true values of the target
variable(s).
You can use as.data.frame to directly access the $data slot. Moreover, all getter
functions for Prediction objects like getPredictionResponse or
getPredictionProbabilities are applicable.
head(as.data.frame(pred))
#> id truth response iter set
#> 1 36 R M 1 test
#> 2 132 M R 1 test
#> 3 145 M R 1 test
#> 4 161 M R 1 test
#> 5 108 M M 1 test
#> 6 178 M M 1 test
head(getPredictionTruth(pred))
#> [1] R M M M M M
#> Levels: M R
head(getPredictionResponse(pred))
#> [1] M R R R M M
#> Levels: M R
The columns iter and set in the data.frame indicate the
resampling iteration and the data set (train or test) for which the prediction was made.
By default, predictions are made for the test sets only.
If predictions for the training set are required, set predict = "train" (for predictions
on the train set only) or predict = "both" (for predictions on both train and test sets)
in makeResampleDesc. In any case, this is necessary for some bootstrap methods
(b632 and b632+) and some examples are shown later on.
Below, we use simple Holdout, i.e., split the data once into a training and test set, as resampling strategy and make predictions on both sets.
## Make predictions on both training and test sets
rdesc = makeResampleDesc("Holdout", predict = "both")
r = resample("classif.lda", iris.task, rdesc, show.info = FALSE)
r
#> Resample Result
#> Task: iris-example
#> Learner: classif.lda
#> Aggr perf: mmce.test.mean=0.0200000
#> Runtime: 0.0176778
r$measures.train
#> iter mmce
#> 1 1 0.02
(Please note that nonetheless the misclassification rate r$aggr is estimated on the test data only.
How to calculate performance measures on the training sets is shown
below.)
A second function to extract predictions from resample results is getRRPredictionList which returns a list of predictions split by data set (train/test) and resampling iteration.
predList = getRRPredictionList(r)
predList
#> $train
#> $train$`1`
#> Prediction: 100 observations
#> predict.type: response
#> threshold:
#> time: 0.00
#> id truth response
#> 123 123 virginica virginica
#> 101 101 virginica virginica
#> 51 51 versicolor versicolor
#> 45 45 setosa setosa
#> 46 46 setosa setosa
#> 3 3 setosa setosa
#> ... (#rows: 100, #cols: 3)
#>
#>
#> $test
#> $test$`1`
#> Prediction: 50 observations
#> predict.type: response
#> threshold:
#> time: 0.00
#> id truth response
#> 109 109 virginica virginica
#> 80 80 versicolor versicolor
#> 40 40 setosa setosa
#> 140 140 virginica virginica
#> 125 125 virginica virginica
#> 10 10 setosa setosa
#> ... (#rows: 50, #cols: 3)
Learner models
In each resampling iteration a Learner is fitted on the respective training set.
By default, the resulting WrappedModels are not included in the
ResampleResult and slot $models is empty.
In order to keep them, set models = TRUE when calling resample, as in the following
survival analysis example.
## 3-fold cross-validation
rdesc = makeResampleDesc("CV", iters = 3)
r = resample("surv.coxph", lung.task, rdesc, show.info = FALSE, models = TRUE)
r$models
#> [[1]]
#> Model for learner.id=surv.coxph; learner.class=surv.coxph
#> Trained on: task.id = lung-example; obs = 112; features = 8
#> Hyperparameters:
#>
#> [[2]]
#> Model for learner.id=surv.coxph; learner.class=surv.coxph
#> Trained on: task.id = lung-example; obs = 111; features = 8
#> Hyperparameters:
#>
#> [[3]]
#> Model for learner.id=surv.coxph; learner.class=surv.coxph
#> Trained on: task.id = lung-example; obs = 111; features = 8
#> Hyperparameters:
The extract option
Keeping complete fitted models can be memory-intensive if these objects are large or
the number of resampling iterations is high.
Alternatively, you can use the extract argument of resample to retain only the
information you need.
To this end you need to pass a function to extract which is applied
to each WrappedModel object fitted in each resampling iteration.
Below, we cluster the mtcars data using the -means algorithm with and keep only the cluster centers.
## 3-fold cross-validation
rdesc = makeResampleDesc("CV", iters = 3)
## Extract the compute cluster centers
r = resample("cluster.kmeans", mtcars.task, rdesc, show.info = FALSE,
centers = 3, extract = function(x) getLearnerModel(x)$centers)
r$extract
#> [[1]]
#> mpg cyl disp hp drat wt qsec
#> 1 16.23333 7.666667 308.9667 214.00000 3.400000 3.564167 16.37000
#> 2 26.00833 4.333333 113.5917 86.08333 4.040833 2.368583 18.88917
#> 3 13.33333 8.000000 444.0000 198.33333 3.003333 4.839667 17.61667
#> vs am gear carb
#> 1 0.1666667 0.3333333 3.666667 3.666667
#> 2 0.8333333 0.6666667 4.083333 1.916667
#> 3 0.0000000 0.0000000 3.000000 3.333333
#>
#> [[2]]
#> mpg cyl disp hp drat wt qsec vs am gear
#> 1 15.5600 8 326.0400 207.00 3.198 3.830000 16.74600 0.000 0.10 3.200
#> 2 26.7125 4 102.8875 86.00 4.145 2.179125 19.05375 0.875 0.75 4.125
#> 3 19.1500 6 174.4000 128.25 3.550 3.136250 17.91000 0.500 0.50 4.000
#> carb
#> 1 3.500
#> 2 1.625
#> 3 3.750
#>
#> [[3]]
#> mpg cyl disp hp drat wt qsec vs
#> 1 25.25000 4 113.6000 82.5000 3.932500 2.622500 19.17000 1.0000000
#> 2 15.12000 8 369.8600 201.9000 3.211000 4.098900 17.05300 0.0000000
#> 3 19.74286 6 183.3143 122.2857 3.585714 3.117143 17.97714 0.5714286
#> am gear carb
#> 1 0.7500000 4.000000 1.500000
#> 2 0.1000000 3.200000 3.200000
#> 3 0.4285714 3.857143 3.428571
As a second example, we extract the variable importances from fitted regression trees using function getFeatureImportance. (For more detailed information on this topic see the feature selection page.)
## Extract the variable importance in a regression tree
r = resample("regr.rpart", bh.task, rdesc, show.info = FALSE, extract = getFeatureImportance)
r$extract
#> [[1]]
#> FeatureImportance:
#> Task: BostonHousing-example
#>
#> Learner: regr.rpart
#> Measure: NA
#> Contrast: NA
#> Aggregation: function (x) x
#> Replace: NA
#> Number of Monte-Carlo iterations: NA
#> Local: FALSE
#> crim zn indus chas nox rm age dis
#> 1 3842.839 952.3849 4443.578 90.63669 3772.273 15853.01 3997.275 3355.651
#> rad tax ptratio b lstat
#> 1 987.4256 568.177 2860.129 0 11255.66
#>
#> [[2]]
#> FeatureImportance:
#> Task: BostonHousing-example
#>
#> Learner: regr.rpart
#> Measure: NA
#> Contrast: NA
#> Aggregation: function (x) x
#> Replace: NA
#> Number of Monte-Carlo iterations: NA
#> Local: FALSE
#> crim zn indus chas nox rm age dis
#> 1 3246.521 3411.444 5806.613 0 2349.776 10125.04 5692.587 2108.059
#> rad tax ptratio b lstat
#> 1 312.6521 2159.42 1104.839 174.6412 15871.53
#>
#> [[3]]
#> FeatureImportance:
#> Task: BostonHousing-example
#>
#> Learner: regr.rpart
#> Measure: NA
#> Contrast: NA
#> Aggregation: function (x) x
#> Replace: NA
#> Number of Monte-Carlo iterations: NA
#> Local: FALSE
#> crim zn indus chas nox rm age dis
#> 1 3785.852 1649.28 4942.119 0 3989.326 18426.87 2604.239 350.8401
#> rad tax ptratio b lstat
#> 1 800.798 2907.556 3871.556 491.6297 12505.88
Stratification and blocking
- Stratification with respect to a categorical variable makes sure that all its values are present in each training and test set in approximately the same proportion as in the original data set. Stratification is possible with regard to categorical target variables (and thus for supervised classification and survival analysis) or categorical explanatory variables.
- Blocking refers to the situation that subsets of observations belong together and must not be separated during resampling. Hence, for one train/test set pair the entire block is either in the training set or in the test set.
Stratification with respect to the target variable(s)
For classification, it is usually desirable to have the same proportion of the classes in
all of the partitions of the original data set.
This is particularly useful in the case of imbalanced classes and small data sets. Otherwise,
it may happen that observations of less frequent classes are missing in some of the training
sets which can decrease the performance of the learner, or lead to model crashes.
In order to conduct stratified resampling, set stratify = TRUE in makeResampleDesc.
## 3-fold cross-validation
rdesc = makeResampleDesc("CV", iters = 3, stratify = TRUE)
r = resample("classif.lda", iris.task, rdesc, show.info = FALSE)
r
#> Resample Result
#> Task: iris-example
#> Learner: classif.lda
#> Aggr perf: mmce.test.mean=0.0132026
#> Runtime: 0.0340035
Stratification is also available for survival tasks. Here the stratification balances the censoring rate.
Stratification with respect to explanatory variables
Sometimes it is required to also stratify on the input data, e.g., to ensure that all
subgroups are represented in all training and test sets.
To stratify on the input columns, specify factor columns of your task data
via stratify.cols.
rdesc = makeResampleDesc("CV", iters = 3, stratify.cols = "chas")
r = resample("regr.rpart", bh.task, rdesc, show.info = FALSE)
r
#> Resample Result
#> Task: BostonHousing-example
#> Learner: regr.rpart
#> Aggr perf: mse.test.mean=21.2385142
#> Runtime: 0.047719
Blocking
If some observations "belong together" and must not be separated when splitting the
data into training and test sets for resampling, you can supply this information via a
blocking factor when creating the task.
## 5 blocks containing 30 observations each
task = makeClassifTask(data = iris, target = "Species", blocking = factor(rep(1:5, each = 30)))
task
#> Supervised task: iris
#> Type: classif
#> Target: Species
#> Observations: 150
#> Features:
#> numerics factors ordered functionals
#> 4 0 0 0
#> Missings: FALSE
#> Has weights: FALSE
#> Has blocking: TRUE
#> Classes: 3
#> setosa versicolor virginica
#> 50 50 50
#> Positive class: NA
Resample descriptions and resample instances
As already mentioned, you can specify a resampling strategy using function makeResampleDesc.
rdesc = makeResampleDesc("CV", iters = 3)
rdesc
#> Resample description: cross-validation with 3 iterations.
#> Predict: test
#> Stratification: FALSE
str(rdesc)
#> List of 4
#> $ id : chr "cross-validation"
#> $ iters : int 3
#> $ predict : chr "test"
#> $ stratify: logi FALSE
#> - attr(*, "class")= chr [1:2] "CVDesc" "ResampleDesc"
str(makeResampleDesc("Subsample", stratify.cols = "chas"))
#> List of 6
#> $ split : num 0.667
#> $ id : chr "subsampling"
#> $ iters : int 30
#> $ predict : chr "test"
#> $ stratify : logi FALSE
#> $ stratify.cols: chr "chas"
#> - attr(*, "class")= chr [1:2] "SubsampleDesc" "ResampleDesc"
The result rdesc inherits from class ResampleDesc (short for
resample description) and, in principle, contains all necessary information about the
resampling strategy including the number of iterations, the proportion of training and test
sets, stratification variables, etc.
Given either the size of the data set at hand or the Task, function makeResampleInstance draws the training and test sets according to the ResampleDesc.
## Create a resample instance based an a task
rin = makeResampleInstance(rdesc, iris.task)
rin
#> Resample instance for 150 cases.
#> Resample description: cross-validation with 3 iterations.
#> Predict: test
#> Stratification: FALSE
str(rin)
#> List of 5
#> $ desc :List of 4
#> ..$ id : chr "cross-validation"
#> ..$ iters : int 3
#> ..$ predict : chr "test"
#> ..$ stratify: logi FALSE
#> ..- attr(*, "class")= chr [1:2] "CVDesc" "ResampleDesc"
#> $ size : int 150
#> $ train.inds:List of 3
#> ..$ : int [1:100] 88 129 94 109 108 43 72 47 137 39 ...
#> ..$ : int [1:100] 129 94 138 83 112 54 29 36 72 137 ...
#> ..$ : int [1:100] 88 138 109 83 112 108 54 29 36 43 ...
#> $ test.inds :List of 3
#> ..$ : int [1:50] 2 5 6 13 14 17 20 21 24 25 ...
#> ..$ : int [1:50] 3 4 7 8 11 12 22 30 34 35 ...
#> ..$ : int [1:50] 1 9 10 15 16 18 19 23 27 28 ...
#> $ group : Factor w/ 0 levels:
#> - attr(*, "class")= chr "ResampleInstance"
## Create a resample instance given the size of the data set
rin = makeResampleInstance(rdesc, size = nrow(iris))
str(rin)
#> List of 5
#> $ desc :List of 4
#> ..$ id : chr "cross-validation"
#> ..$ iters : int 3
#> ..$ predict : chr "test"
#> ..$ stratify: logi FALSE
#> ..- attr(*, "class")= chr [1:2] "CVDesc" "ResampleDesc"
#> $ size : int 150
#> $ train.inds:List of 3
#> ..$ : int [1:100] 149 58 120 44 148 29 66 46 124 137 ...
#> ..$ : int [1:100] 51 58 64 148 56 46 124 8 14 137 ...
#> ..$ : int [1:100] 149 51 120 44 64 56 29 66 8 14 ...
#> $ test.inds :List of 3
#> ..$ : int [1:50] 3 8 12 14 17 22 23 24 32 34 ...
#> ..$ : int [1:50] 1 2 4 6 10 11 13 26 29 30 ...
#> ..$ : int [1:50] 5 7 9 15 16 18 19 20 21 25 ...
#> $ group : Factor w/ 0 levels:
#> - attr(*, "class")= chr "ResampleInstance"
## Access the indices of the training observations in iteration 3
rin$train.inds[[3]]
#> [1] 149 51 120 44 64 56 29 66 8 14 83 65 97 114 13 3 104
#> [18] 88 130 81 89 23 63 131 92 31 41 78 72 139 67 10 57 12
#> [35] 107 74 70 116 36 24 35 93 126 111 75 91 80 85 42 30 22
#> [52] 1 69 113 87 26 17 150 119 4 138 129 147 38 99 60 142 50
#> [69] 122 40 127 43 96 34 141 106 79 133 145 125 135 108 52 109 37
#> [86] 61 84 59 39 82 32 53 94 6 45 86 95 2 68 11
The result rin inherits from class ResampleInstance and contains
lists of index vectors for the train and test sets.
If a ResampleDesc is passed to resample, it is instantiated internally. Naturally, it is also possible to pass a ResampleInstance directly.
While the separation between resample descriptions, resample instances, and the resample function itself seems overly complicated, it has several advantages:
- Resample instances readily allow for paired experiments, that is comparing the performance of several learners on exactly the same training and test sets. This is particularly useful if you want to add another method to a comparison experiment you already did. Moreover, you can store the resample instance along with your data in order to be able to reproduce your results later on.
rdesc = makeResampleDesc("CV", iters = 3)
rin = makeResampleInstance(rdesc, task = iris.task)
## Calculate the performance of two learners based on the same resample instance
r.lda = resample("classif.lda", iris.task, rin, show.info = FALSE)
r.rpart = resample("classif.rpart", iris.task, rin, show.info = FALSE)
r.lda$aggr
#> mmce.test.mean
#> 0.02
r.rpart$aggr
#> mmce.test.mean
#> 0.05333333
- In order to add further resampling methods you can simply derive from the ResampleDesc and ResampleInstance classes, but you do neither have to touch resample nor any further methods that use the resampling strategy.
Usually, when calling makeResampleInstance the train and test index sets are drawn randomly. Mainly for holdout (test sample) estimation you might want full control about the training and tests set and specify them manually. This can be done using function makeFixedHoldoutInstance.
rin = makeFixedHoldoutInstance(train.inds = 1:100, test.inds = 101:150, size = 150)
rin
#> Resample instance for 150 cases.
#> Resample description: holdout with 0.67 split rate.
#> Predict: test
#> Stratification: FALSE
Aggregating performance values
In each resampling iteration we get performance values (for each measure we wish to calculate), which are then aggregated to an overall performance.
For the great majority of common resampling strategies (like holdout, cross-validation, subsampling) performance values are calculated on the test data sets only and for most measures aggregated by taking the mean (test.mean).
Each performance Measure in mlr has a corresponding default aggregation
method which is stored in slot $aggr.
The default aggregation for most measures is test.mean.
One exception is the root mean square error (rmse).
## Mean misclassification error
mmce$aggr
#> Aggregation function: test.mean
mmce$aggr$fun
#> function (task, perf.test, perf.train, measure, group, pred)
#> mean(perf.test)
#> <bytecode: 0xc6e6978>
#> <environment: namespace:mlr>
## Root mean square error
rmse$aggr
#> Aggregation function: test.rmse
rmse$aggr$fun
#> function (task, perf.test, perf.train, measure, group, pred)
#> sqrt(mean(perf.test^2))
#> <bytecode: 0x19156600>
#> <environment: namespace:mlr>
You can change the aggregation method of a Measure via function setAggregation. All available aggregation schemes are listed on the aggregations documentation page.
Example: One measure with different aggregations
The aggregation schemes test.median, test.min, and test.max compute the median, minimum, and maximum of the performance values on the test sets.
mseTestMedian = setAggregation(mse, test.median)
mseTestMin = setAggregation(mse, test.min)
mseTestMax = setAggregation(mse, test.max)
mseTestMedian
#> Name: Mean of squared errors
#> Performance measure: mse
#> Properties: regr,req.pred,req.truth
#> Minimize: TRUE
#> Best: 0; Worst: Inf
#> Aggregated by: test.median
#> Arguments:
#> Note: Defined as: mean((response - truth)^2)
rdesc = makeResampleDesc("CV", iters = 3)
r = resample("regr.lm", bh.task, rdesc, measures = list(mse, mseTestMedian, mseTestMin, mseTestMax))
#> Resampling: cross-validation
#> Measures: mse mse mse mse
#> [Resample] iter 1: 28.164474328.164474328.164474328.1644743
#> [Resample] iter 2: 17.593981817.593981817.593981817.5939818
#> [Resample] iter 3: 24.957218724.957218724.957218724.9572187
#>
#> Aggregated Result: mse.test.mean=23.5718916,mse.test.median=24.9572187,mse.test.min=17.5939818,mse.test.max=28.1644743
#>
r
#> Resample Result
#> Task: BostonHousing-example
#> Learner: regr.lm
#> Aggr perf: mse.test.mean=23.5718916,mse.test.median=24.9572187,mse.test.min=17.5939818,mse.test.max=28.1644743
#> Runtime: 0.0496087
r$aggr
#> mse.test.mean mse.test.median mse.test.min mse.test.max
#> 23.57189 24.95722 17.59398 28.16447
Example: Calculating the training error
Below we calculate the mean misclassification error (mmce) on the training
and the test data sets. Note that we have to set predict = "both" when calling makeResampleDesc
in order to get predictions on both training and test sets.
mmceTrainMean = setAggregation(mmce, train.mean)
rdesc = makeResampleDesc("CV", iters = 3, predict = "both")
r = resample("classif.rpart", iris.task, rdesc, measures = list(mmce, mmceTrainMean))
#> Resampling: cross-validation
#> Measures: mmce.train mmce.test
#> [Resample] iter 1: 0.0200000 0.1000000
#> [Resample] iter 2: 0.0400000 0.0400000
#> [Resample] iter 3: 0.0400000 0.0400000
#>
#> Aggregated Result: mmce.test.mean=0.0600000,mmce.train.mean=0.0333333
#>
r$measures.train
#> iter mmce mmce
#> 1 1 0.02 0.02
#> 2 2 0.04 0.04
#> 3 3 0.04 0.04
r$aggr
#> mmce.test.mean mmce.train.mean
#> 0.06000000 0.03333333
Example: Bootstrap
In out-of-bag bootstrap estimation new data sets are drawn from the data set with replacement, each of the same size as . In the -th iteration, forms the training set, while the remaining elements from , i.e., , form the test set.
The b632 and b632+ variants calculate a convex combination of the training performance and the out-of-bag bootstrap performance and thus require predictions on the training sets and an appropriate aggregation strategy.
## Use bootstrap as resampling strategy and predict on both train and test sets
rdesc = makeResampleDesc("Bootstrap", predict = "both", iters = 10)
## Set aggregation schemes for b632 and b632+ bootstrap
mmceB632 = setAggregation(mmce, b632)
mmceB632plus = setAggregation(mmce, b632plus)
mmceB632
#> Name: Mean misclassification error
#> Performance measure: mmce
#> Properties: classif,classif.multi,req.pred,req.truth
#> Minimize: TRUE
#> Best: 0; Worst: 1
#> Aggregated by: b632
#> Arguments:
#> Note: Defined as: mean(response != truth)
r = resample("classif.rpart", iris.task, rdesc, measures = list(mmce, mmceB632, mmceB632plus),
show.info = FALSE)
head(r$measures.train)
#> iter mmce mmce mmce
#> 1 1 0.04000000 0.04000000 0.04000000
#> 2 2 0.04000000 0.04000000 0.04000000
#> 3 3 0.01333333 0.01333333 0.01333333
#> 4 4 0.02666667 0.02666667 0.02666667
#> 5 5 0.01333333 0.01333333 0.01333333
#> 6 6 0.02000000 0.02000000 0.02000000
## Compare misclassification rates for out-of-bag, b632, and b632+ bootstrap
r$aggr
#> mmce.test.mean mmce.b632 mmce.b632plus
#> 0.05804883 0.04797219 0.04860054
Convenience functions
The functionality described on this page allows for much control and flexibility. However, when quickly trying out some learners, it can get tedious to type all the code for defining the resampling strategy, setting the aggregation scheme and so on. As mentioned above, mlr includes some pre-defined resample description objects for frequently used strategies like, e.g., 5-fold cross-validation (cv5). Moreover, mlr provides special functions for the most common resampling methods, for example holdout, crossval, or bootstrapB632.
crossval("classif.lda", iris.task, iters = 3, measures = list(mmce, ber))
#> Resampling: cross-validation
#> Measures: mmce ber
#> [Resample] iter 1: 0.0200000 0.0158730
#> [Resample] iter 2: 0.0400000 0.0415140
#> [Resample] iter 3: 0.0000000 0.0000000
#>
#> Aggregated Result: mmce.test.mean=0.0200000,ber.test.mean=0.0191290
#>
#> Resample Result
#> Task: iris-example
#> Learner: classif.lda
#> Aggr perf: mmce.test.mean=0.0200000,ber.test.mean=0.0191290
#> Runtime: 0.0388408
bootstrapB632plus("regr.lm", bh.task, iters = 3, measures = list(mse, mae))
#> Resampling: OOB bootstrapping
#> Measures: mse.train mae.train mse.test mae.test
#> [Resample] iter 1: 18.9037446 3.0912153 29.2662169 3.7698624
#> [Resample] iter 2: 17.9389954 3.0343581 26.3888260 3.5992878
#> [Resample] iter 3: 20.9092738 3.2640991 23.7739540 3.6788560
#>
#> Aggregated Result: mse.b632plus=23.9312510,mae.b632plus=3.4912886
#>
#> Resample Result
#> Task: BostonHousing-example
#> Learner: regr.lm
#> Aggr perf: mse.b632plus=23.9312510,mae.b632plus=3.4912886
#> Runtime: 0.0676453